SUB DISTRIBUTED BIOINFORMATICS CENTRE
The Bioinformatics Centre a Sub Centre -Distributed Information Centre (Sub-DIC) at Bharathidasan University is an offshoot of National Facility for Marine Cyanobacteria (NFMC) sponsored by DBT Govt of IndiaOpen Source Bioinformatics Tools
|
|
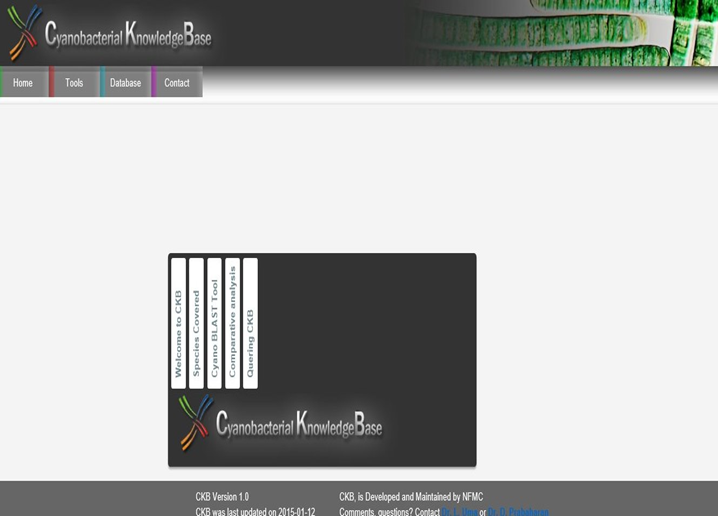
Cyanobacterrial Knowledge Base (CKB) ,Cyanobacterial KnowldegeBase (CKB) was the first of its kind developed to provide wide opportunities for understanding the metabolic organization and support computational research with the availability of completely sequenced cyanobacterial genomes. The database contains tools for sequence analysis and the species report and the gene report which provides the details about each species and gene (including sequence features and gene ontology annotations) respectively. The database also includes cyanoBLAST, an advanced tool that facilitates comparative analysis, among cyanobacterial genomes and genomes of E. coli (prokaryote) and Arabidopsis (eukaryote). |
|
|
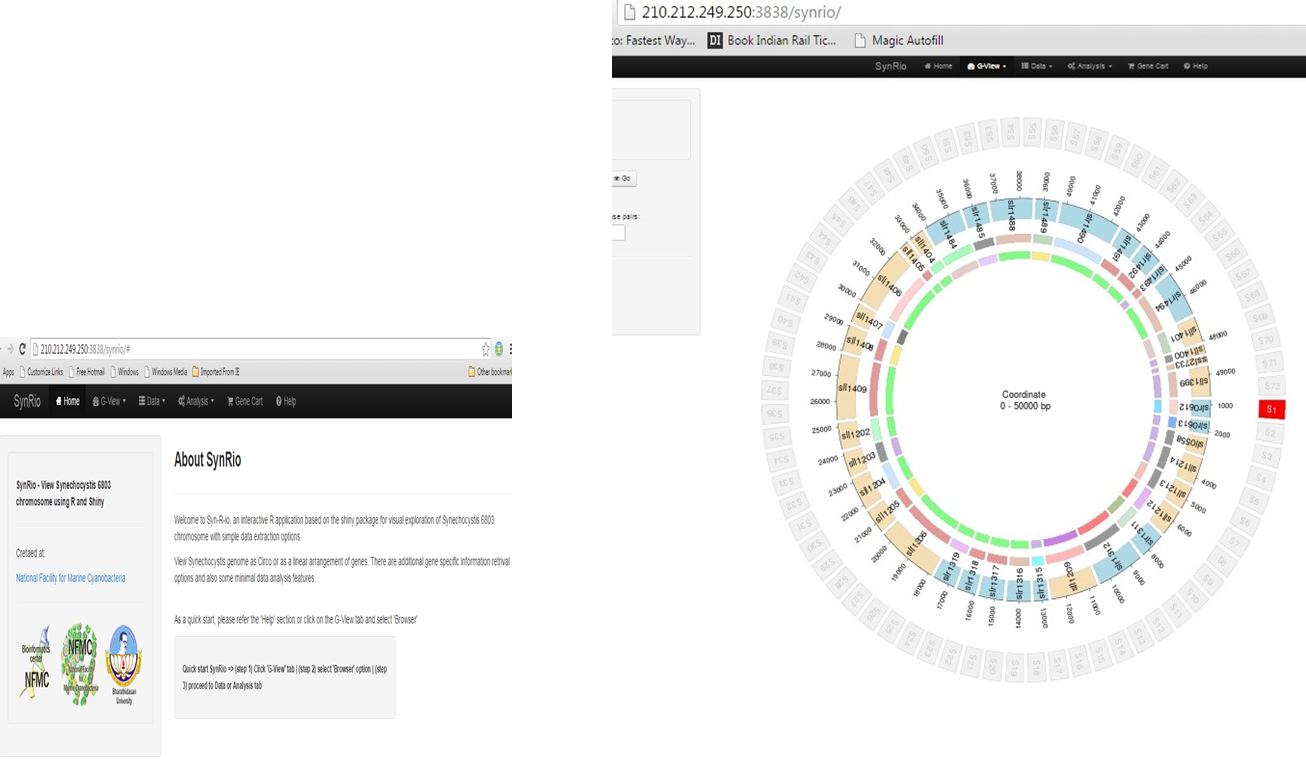
Syn - R - io ,an open source cyanobacterial model organism genomics and gene analysis platform. The model organism Synechocystis PCC 6803 genome sequence has been used as a starting point and all the genome and gene based annotations were gathered and compiled to visually query and analyse the organism. The platform is created entirely using R programming language with Shiny package and server. |
|
|
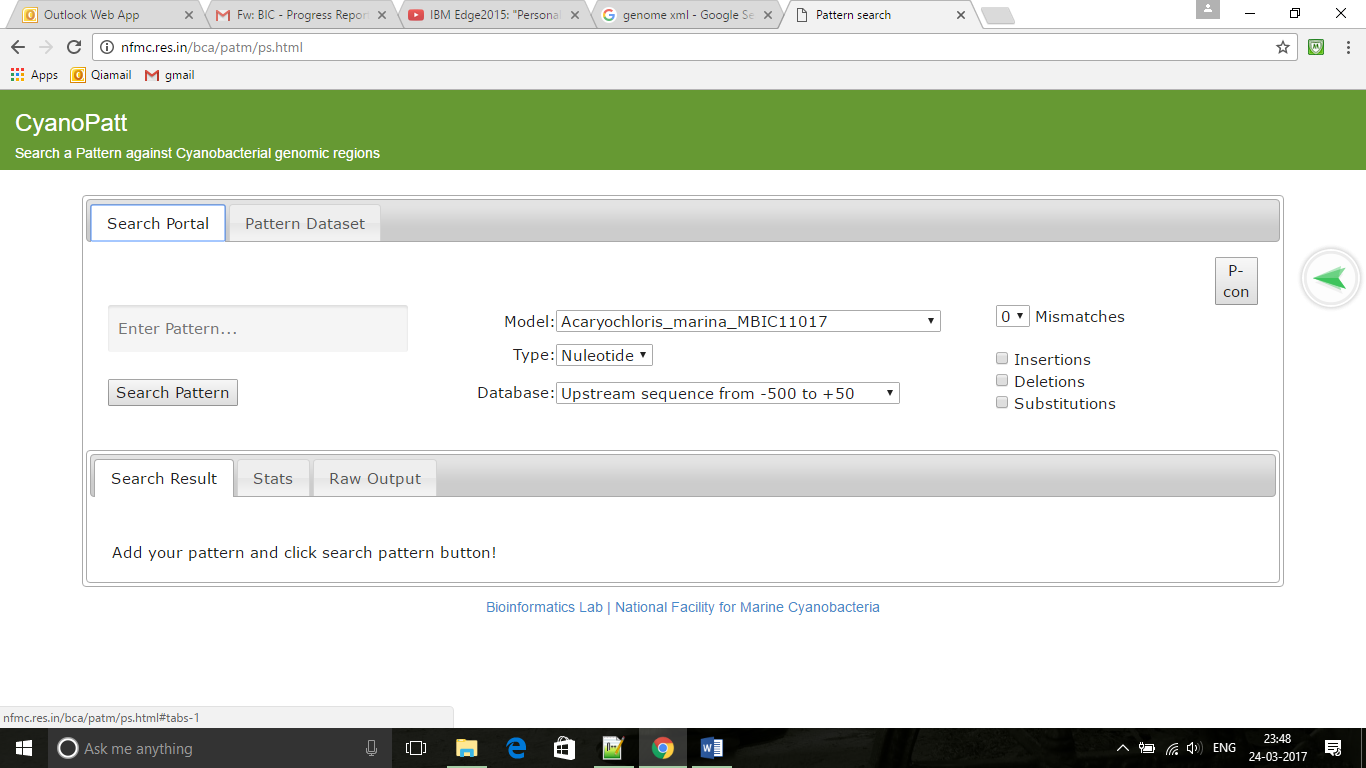
Cyanopatt ,The cis and trans regulatory mechanisms related to cyanobacterial genome is an interesting area to venture to better comment on the signalling mechanism which are of economic importance. We attempt to put together data pertaining to regulatory regions of 74 cyanobacterial genomes and manually curated information on transcription factors and there experimentally proven binding sites in the promoter region. Cyanopatt is also helpful in predicting the probable transcription factor binding sites in interested cyano genome. |